Who: Dr. Sandy Martin, Professor
Department of Cell and Developmental Biology
What: 2019 Fall Graduate Student Seminar
When: Monday, Oct. 18th at Noon
Where: SI 1086
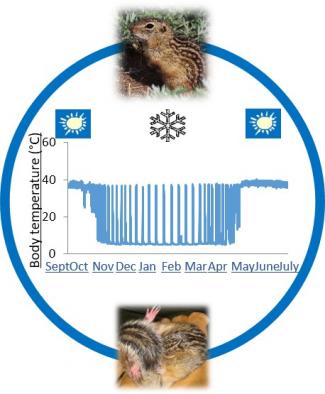
If many phylogenetically interspersed species of mammals can safely lower their metabolic rate to a few percent of resting, and their body temperature to near freezing, why can’t humans? Engineering a reversible metabolic depression in our species that is based on natural hibernation would be a huge boon for the treatment of cardiac arrest, stroke and trauma, routine surgery and manned space missions. The goal of our ongoing work is to exploit the natural rhythms of hibernation to elucidate the biochemical pathways that make this extraordinary phenotype possible. Small-bodied hibernators, including the 13-lined ground squirrel (Ictidomys tridecemlineatus), exploit winter’s cold to save energy by entering a state called torpor. Torpor is discontinuous, giving rise to a cyclical pattern of metabolic depression and activation with a period of about two weeks. Because torpor, and thus the cycles between torpor and arousal that comprise hibernation, only occurs during part of the year (fall and winter), hibernation can be thought of as a cycle within a cycle, ie a torpor-arousal cycle that is embedded within the inactive phase of an annual cycle between activity and inactivity. This dynamic and extreme circannual phenotype for a mammal, like all phenotypes, must be orchestrated via gene expression programs and is thought to include not only the mechanisms that underlie the hibernator’s exceptional metabolic flexibility, but also mechanisms that confer tissue protection from damage by cold and ischemia followed by rapid reperfusion. We hypothesize that differential gene expression patterns control the tissue remodeling needed to orchestrate and survive torpor; this predicts that key genes can be identified by analyzing differential gene expression in carefully timed and collected samples representing the distinct phases of the hibernator’s phenotype. Current work is focused on the analyses of RNA-seq data, including for differential expression, splicing and modification as a function of hibernation physiology. The utility and limitations of these and other –omic datasets in understanding hibernation will be discussed.
Everyone is welcome to the seminar. If you would like to meet with the speaker, please email Dr. Greg Ragland.